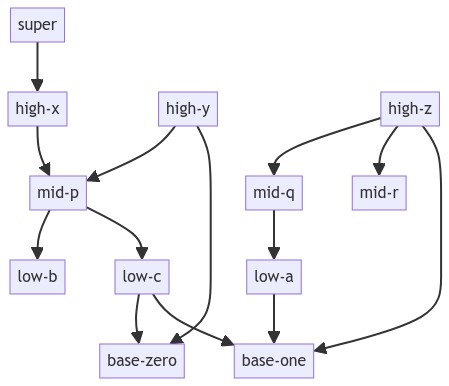
If you look at this DAG (directed acyclic graph):
I want to create a dict which maps the distance from the lowest node(s) to all others nodes which is similar to the x position (height) of each node from the bottom in the rendered graph. For that given graph it would be:
distance_nodes_map: {
0: {'base-zero', 'base-one'},
1: {'low-b', 'low-a', 'low-c'},
3: {'high-x', 'high-z', 'high-y'},
2: {'mid-r', 'mid-q', 'mid-p'},
4: {'super'}
}
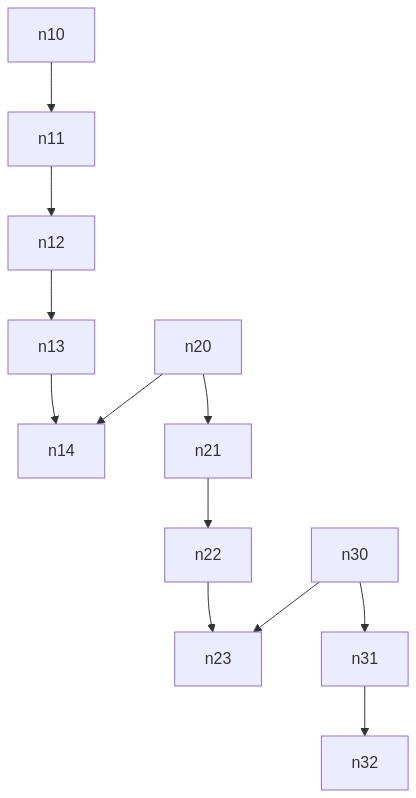
I wrote an algorithm which worked for that graph above but then I've tested another graph and it didn't work anymore. I tried some algorithms and functions like shortest path or descendants_at_distance but I don't think they are really helpful as an input to calculate the distances.
My algorithm doesn't work for instance for this graph:
https://gist.github.com/timaschew/3b08a07243fa6f43773014ef5e705c96
Here is gist which contains:
- a python script which reads a YAML file, the dependency/graph structure and generates a HTML with a rendered mermaid graph (I've removed my algorithm to calculate the distances in a wrong way)
- both graphs which are shown here, as a YAML file
from How to calculate overall distances from lowest root(s) of a directed graph with networkx
No comments:
Post a Comment